
Biosynthetic pathway Pantaphos a natural product phosphonate
Analysis of the biosynthetic pathway of Pantaphos, a natural product of phosphonate, and discovery of methyltransferase with new functions
Abstract
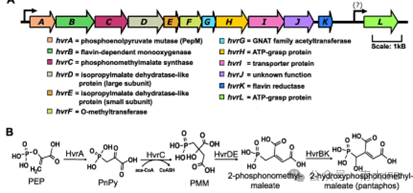
pantaphos, a natural product of phosphonate, is a key pathogenic factor of onion bulb rot produced by Pantoea ananatis, a plant pathogen. Its biosynthetic gene cluster starts from pepM and contains 11 genes in total. It’s called Hivir (High Virulence).
Natural products of phosphonates can simulate phosphate esters of biomolecules, interfere with cellular metabolic processes and signal transduction, and thus show antibacterial, anticancer and herbicidal activities, so they have attracted wide attention.
Recently, Professor William Metcalf’s team at the University of Illinois at Urbana-Champaign clarified the complete biosynthesis of pantaphos through bioinformatics analysis, enzyme reactions in vitro, and allogenic expression in vivo. A self-detoxifying methyltransferase and a flavin monooxygenase with flavin reductase activity revealed. The results were published in Angew.Chem.Int.Ed. A magazine.
content
Previous studies showed that the first 6 genes on Hivir cluster hvrA (PEP mutase), hvrB (FAD-dependent monooxase), hvrC (phosphonomethylmalate), PMM synthase, hvrDE (size subunit of cis aconitase) and hvrF (methyltransferase) highly correlated with onion lesions caused by Panthenia.
Among them, hvrC-E is a homologous gene of tricarboxylic acid cycle and leucine biosynthetase. Based on this, the authors hypothesized a pantaphos biosynthetic pathway that follows similar chemical processes to the tricarboxylic acid cycle and leucine biosynthesis, but does not involve the hvrF gene (Figure 1).
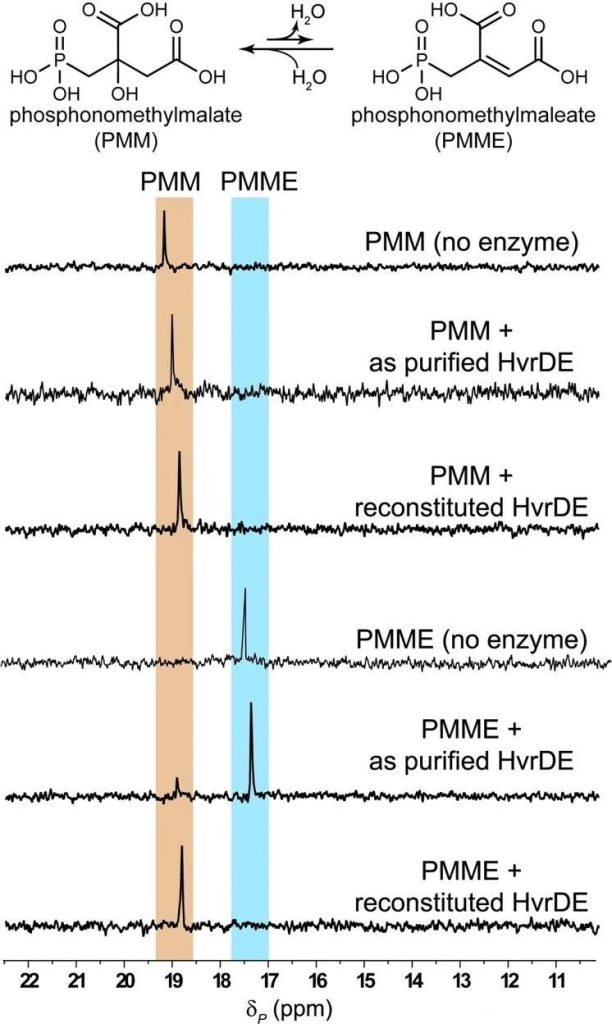
The authors first attempt to characterize the functions of HvrDE. He performed the enzyme reaction using PMM as a substrate and did not detect dehydration products, while using PMME, a product of HvrDE on the presumed path, as a substrate, a small amount of hydration products could be detected (Figure 2).
The authors speculate that the dehydration reaction catalyzed by HvrDE is thermodynamically unfavorable and may require downstream enzymes to consume the product to drive the dehydration reaction.
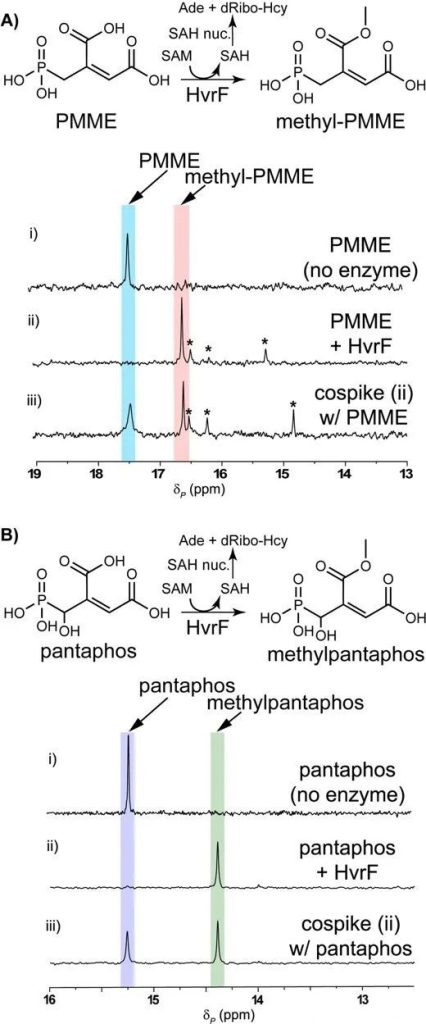
Based on the presumed biosynthesis process, the authors added HvrBK and found that PMME not hydroxylated. As A result, the authors reacted HvrF, a functional methyltransferase, with PMME and found that methylated PMME accumulated (Figure 3A).
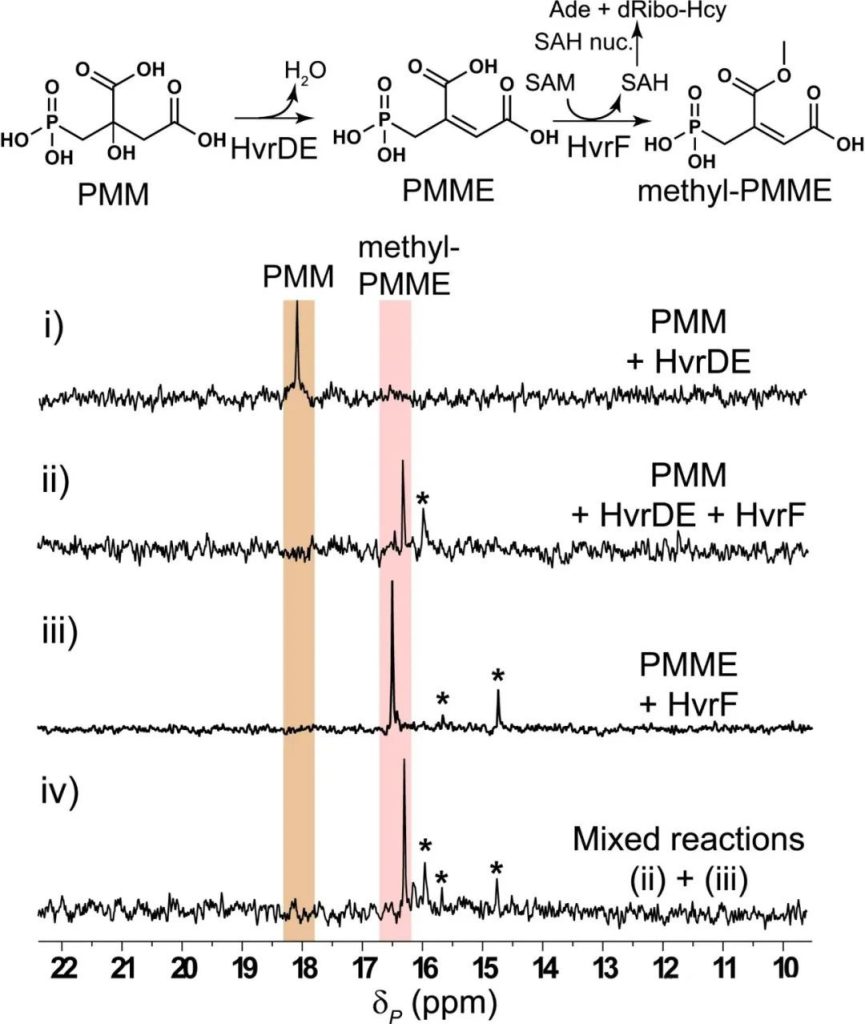
Next, the authors tried a one-pot enzyme method, using PMM as substrate, adding HvrDE and HvrF and corresponding cofactors, and successfully detected methylated PMME, confirming the previous conjconjation that downstream methyltransferase HvrF catalyzed PMME methylation and thus drove upstream HvrDE to catalyze PMM dehydration to form PMME (Figure 4).
Moreover, the authors found that HvrF had a certain substrate breadth and could also catalyze the methylation of pantaphos (Figure 3b). The authors speculated that methylation of pantaphos and its intermediates might be a self-resistant measure taken by the strain.
Subsequently, the author used methylated PMME as substrate to react with HvrBK and detected the hydroxylation product, namely methylated pantaphos, further proving that methylated PMME is an intermediate in the biosynthesis of pantaphos (Figure 5).
However, the authors found that the product could detected even without adding HvrK (flavin reductase) in the enzyme reaction, indicating that HvrB itself has flavin reductase activity, and the enzyme activity experiment showed that the reduction efficiency of HvrB far lower than that of HvrK.
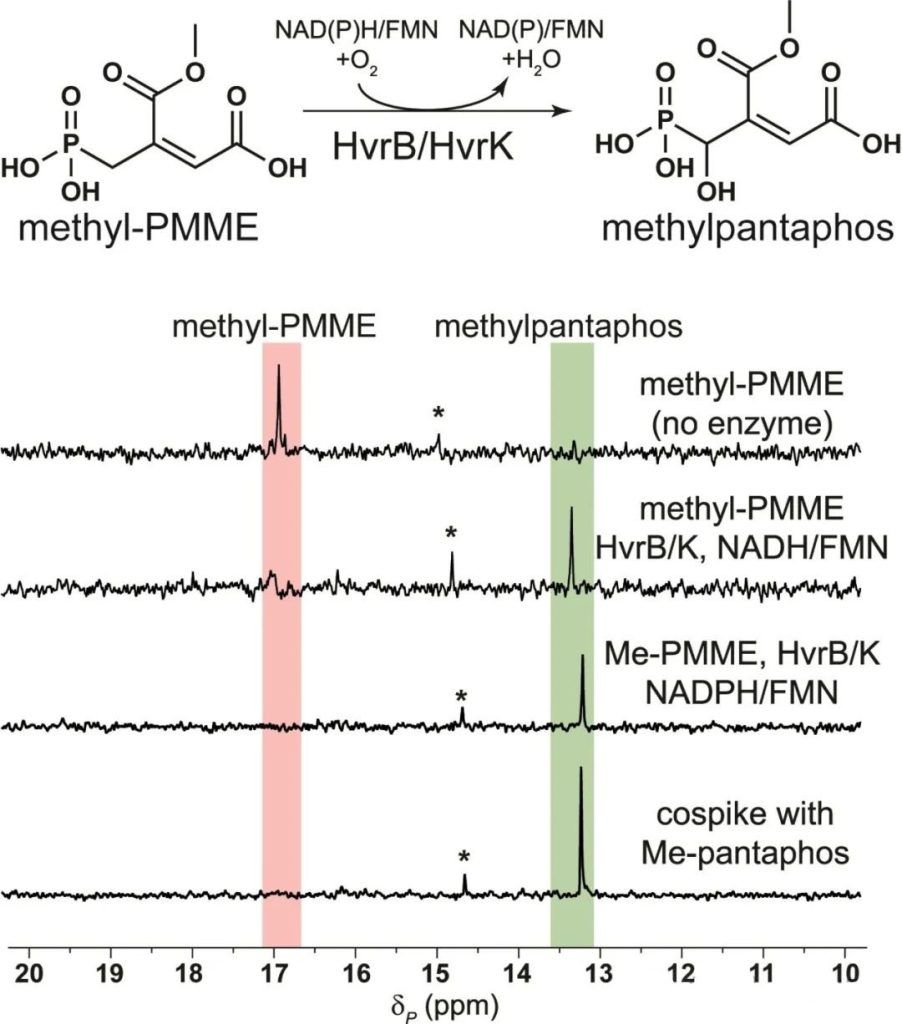
In addition, the authors found that methylated pantaphos would slowly and spontaneously hydrolyze to form pantaphos in the absence of enzymes.
The authors suggested that there might a hydrolase in the strain to accelerate the process, but no possible hydrolase found after functional verification of other genes on the cluster.
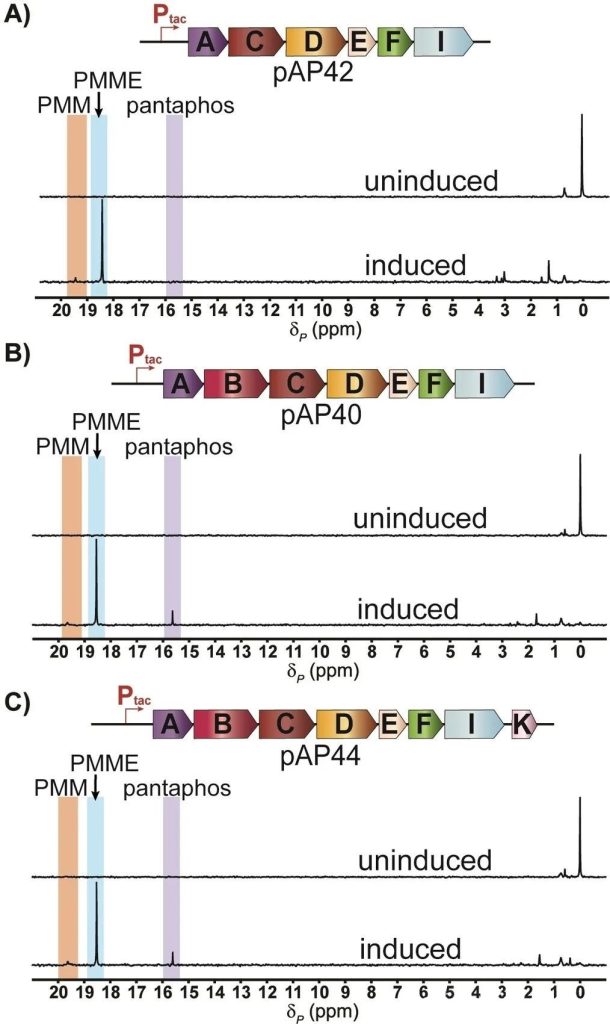
So far, the authors have found all the genes needed to produce pantaphos by enzyme reaction in vitro. Heterologous expression of this minimal gene combination in Escherichia coli also successfully accumulated pantaphos (Figure 6).
Summary
In summary, the authors analyzed the complete biosynthesis process of pantaphos through enzyme reaction in vitro and heterologous expression in vivo, and revealed a methyltransferase that may have self-detoxification function, providing a certain reference for subsequent plant disease control.